PhyML, software developed in Montpellier to track the Covid-19 epidemic
The PhyML bioinformatics software, developed over the past fifteen years at the Montpellier Laboratory of Computer Science, Robotics and Microelectronics (Lirmm) and made available to the international scientific community, is now an important tool for tracing the origins of the Covid-19 pandemic and monitoring the evolution of the virus.
With the emergence of SARS-CoV-2, the virus responsible for COVID-19, the PhyML software, already used to track more traditional viral epidemics such as seasonal flu, is now running at full capacity. "Weare working hard, the software is constantly evolving, we are implementing algorithms to go faster and process more and more data sets, developing new features..." explains Stéphane Guindon, CNRS researcher at the Montpellier Laboratory of Computer Science, Robotics and Microelectronics (Lirmm).
Developed during his thesis in 2003 by this bioinformatician and his thesis supervisor, Olivier Gascuel, PhyML was one of the first free software programs available via the bioinformatics platform hosted by Lirmm. "Wemake our servers available to research laboratories because these phylogenetic analyses are very computationally intensive. Some can take hours, days, or even weeks." Each year, the ATGC platform performs approximately 350,000 hours of computing for laboratories located in France, but also and above all throughout Europe, China, the United States, and elsewhere.
Tracing a chain of transmission
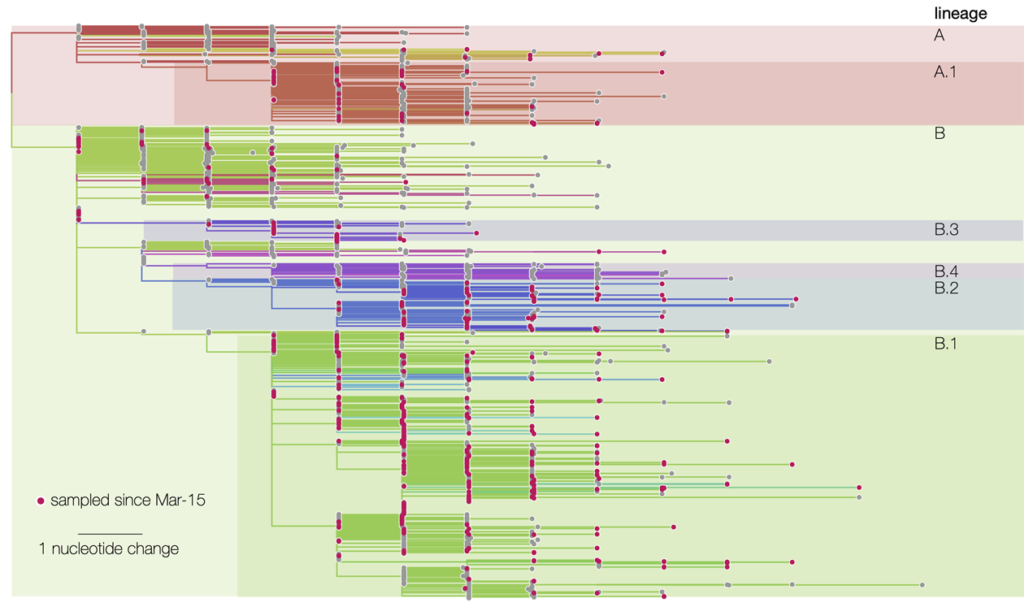
But what is PhyML used for, and why is it so useful during an epidemic? Because this bioinformatics software is capable of tracing the genealogy, or phylogeny, of any group of organisms with DNA. "Aphylogenetic tree is a bit like a family tree, which allows us to trace the family relationships between individuals in the same family. This 'genealogy' is reconstructed here by comparing the genomes of different species. In the case of a virus, we analyze the ~30,000 nucleotides that make up its genome in order to trace a chain of transmission."
SARS-CoV-2 DNA sequences were compared by the software with all those contained in existing databases, establishing a strong link with a virus present in bats and pangolins. "Thisvirus circulates in bats but does not cause mortality in this species, unlike what we are currently seeing in humans, "notes Stéphane Guindon. Analysis of these sequences also determined that the current epidemic originated from a single zoonotic event, in other words, a single case of animal-to-human transmission."If there had been several events, the virus tree would not have the same shape; it would have several sub-trees, each corresponding to a transfer of a viral strain from animals to humans."
Monitoring and predicting the evolution of the virus
Phylogeny has helped answer other important questions about the epidemic. "From the beginning of March, the first phylogenetic analyses showed us that the virus population size was doubling every 5 to 7 days," says the researcher. Phylogenetic analyses using PhyML could also help to better predict the long-term evolution of SARS-CoV-2. For example, every year phylogenetics helps to determine which strains of the influenza virus are most likely to develop the following winter, thereby contributing to the design of effective vaccines.