NUMEV Seminar “Data-driven modeling and time series prediction using the generalized Langevin equation”
This event has passed!
NUMEV seminars are open to a wide audience of students and researchers from all disciplines who wish to learn more about the current research areas of the NUMEV-MIPS community (Mathematics, Computer Science, Physics, and Systems) or about opportunities to develop their skills and expertise.
By Roland Netz, Department of Physics, Free University Berlin
Most systems of scientific interest are N-body interacting systems. Their kinetics are generally described in terms of low-dimensional reaction coordinates, which are usually influenced by the entire system. The dynamics of such a reaction coordinate are governed by the generalized Langevin equation (GLE), a stochastic integro-differential equation, and involve a memory function, which describes how these dynamics depend on previous values of the reaction coordinate. The GLE is therefore a systematic non-Markovian description of the dynamics of a system in terms of coarse-grained variables. We have recently introduced a new hybrid projection scheme that allows GLE parameters to be extracted from time series data in a form suitable for analytical and numerical processing. In this talk, I will discuss some examples in which GLE can be used to interpret and model data in different scientific fields.
Data-based modeling and time series prediction using the generalized Langevin equation
Roland Netz, Department of Physics, Free University Berlin
Abstract
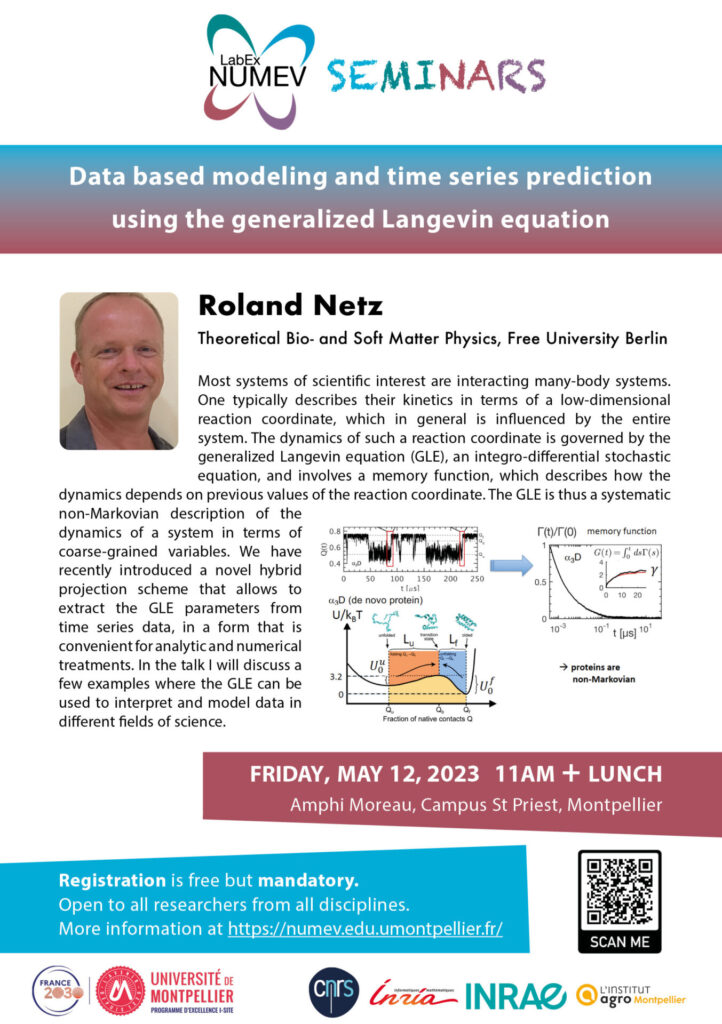
Most systems of scientific interest are interacting many-body systems. One typically describes their kinetics in terms of a low-dimensional reaction coordinate, which in general is influenced by the entire system. The dynamics of such a reaction coordinate is governed by the generalized Langevin equation (GLE), an integro-differential stochastic equation, and involves a memory function, which describes how the dynamics depends on previous values of the reaction coordinate. The GLE is thus a systematic non-Markovian description of the dynamics of a system in terms of coarse-grained variables. We have recently introduced a novel hybrid projection scheme that allows the GLE parameters to be extracted from time series data in a form that is convenient for analytical and numerical treatments. In the talk, I discuss a few examples where the GLE can be used to interpret and model data in different fields of science.
Protein-folding kinetics is typically described as Markovian (i.e., memoryless) diffusion in a one-dimensional free energy landscape, governed by an instantaneous friction coefficient that is fitted to reproduce experimental or simulated folding times. According to this view, the folding time is dominated by the exponential of the folding free energy barrier, the Arrhenius factor, where the friction coefficient only sets the pre-exponential time scale and plays a subordinate role. By analyzing large-scale molecular dynamics simulation trajectories of fast-folding proteins from the Shaw group using the special-purpose computer ANTON, it is demonstrated that the friction characterizing protein folding exhibits significant memory with a decay time that is of the same order as the folding and unfolding times. Non-Markovian modeling not only reproduces simulations accurately but also demonstrates that memory friction effects lead to anomalous and drastically modified protein kinetics. For the set of proteins for which simulations are available, it is shown that the folding and unfolding times are not dominated by the free-energy barrier but rather by the non-Markovian friction.
Memory effects are also present for non-equilibrium systems. Using an appropriate non-equilibrium formulation of the GLE, it is demonstrated that the motion of living organisms is characterized by memory friction, which allows to characterize internal feedback loops of such organisms and to classify and sort individual organisms. The GLE can even be used to predict complex phenomena such as weather data.
Receive a weekly summary of the UM agenda
* By entering your email address, you agree to receive a weekly summary of the UM calendar by email and acknowledge that you have read ourprivacy policy. You can unsubscribe at any time using the unsubscribe linkor by contacting us by email.
